Theme 2 : Regulation of mRNA stability by small regulatory RNAs in Bacillus subtilis
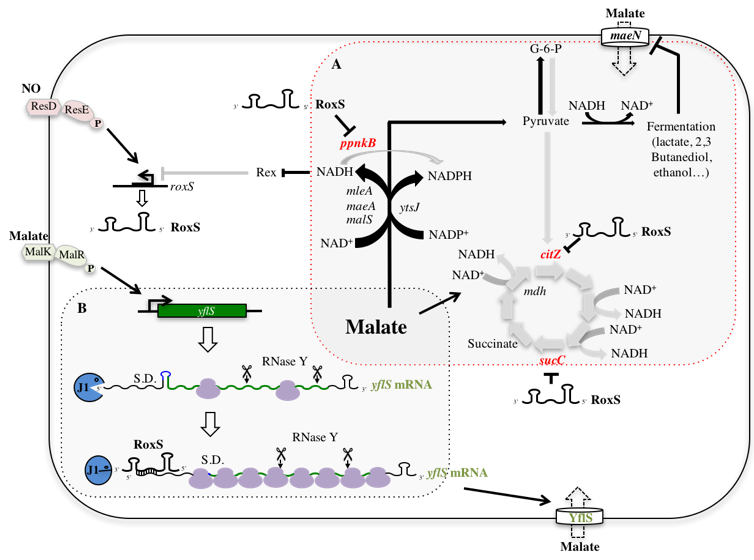
Transcriptional studies have shown that Gram-positive bacteria have about a hundred small regulatory RNAs (sRNAs). However, very few of these sRNAs have been characterized in B. subtilis. As with RNA degradation studies, non-coding RNAs have mainly been studied in Gram-negative bacteria such as E. coli and its relatives. Most sRNAs studied affect translation of mRNAs by hybridizing with the ribosome binding region. However, recent studies have shown that sRNAs can modify the stability of certain RNAs without affecting their translation, by recruiting or directly interfering with the RNA degradation machinery of the bacterium. In the model organism of Gram-positive bacteria, B. subtilis, only a few non-coding RNAs have been characterized and their regulatory mechanisms remain largely unknown. From the few studies carried out up to now, it is clear that the regulatory mechanisms by the non-coding RNAs in the Firmicutes are different from those found in the Proteobacteria. Indeed, the Hfq protein, which plays a key role in to promote interactions between non-in RNA and target mRNA in Gram-negatives, is not required in B. subtilis. In addition, RNase E, responsible for the degradation of target mRNAs after pairing with their regulatory RNAs in E. coli, does not exist in B. subtilis.To better understand the how regulatory RNAs operate in B. subtilis, we hare interested in characterizing several examples of sRNAs and how they interact with the degradation machinery. In this context, we recently characterized the RoxS regulatory RNA, which is conserved in the pathogenic bacterium S. aureus (where it is called RsaE). We showed that the transcription of this RNA is directly regulated by 2 transcription factors: 1. The ResD-ResE two-component system, which responds to the presence of nitric oxide (NO) or low oxygen levels in the medium, and activates the RoxS transcription. 2. The Rex transcription inhibitor, which reacts to the NAD:NADH ratio in the cell. We showed that RoxS impacts the expression of more than 35 mRNAs in the cell, the majority of which are related to the oxidative stress response. Several mRNAs in the Krebs cycle are also regulated by RoxS, including the mRNA encoding succinate dehydrogenase (sucCD). In this case, RoxS pairs with the ribosome binding region to block translation initiation. We have also shown that RoxS is able to directly interfere with the degradation of yflS mRNA, encoding a malate transporter. Indeed, the pairing of RoxS with the 5’ end of this mRNA prevents the exoribonuclease RNase J1 from degrading it.